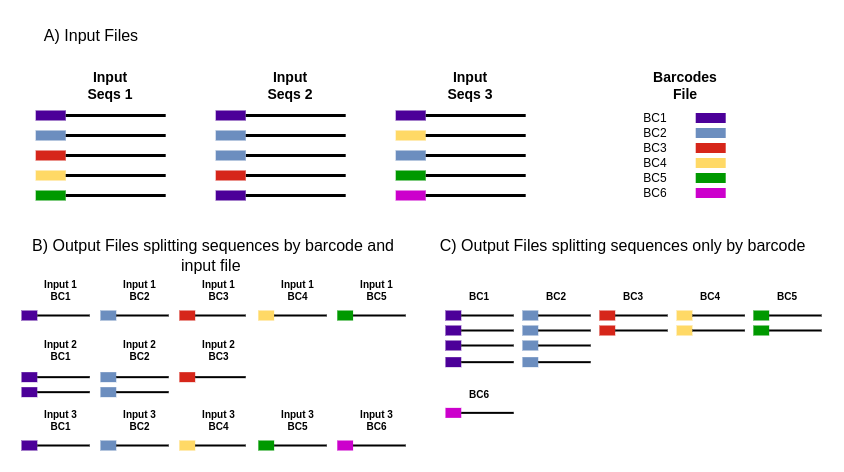
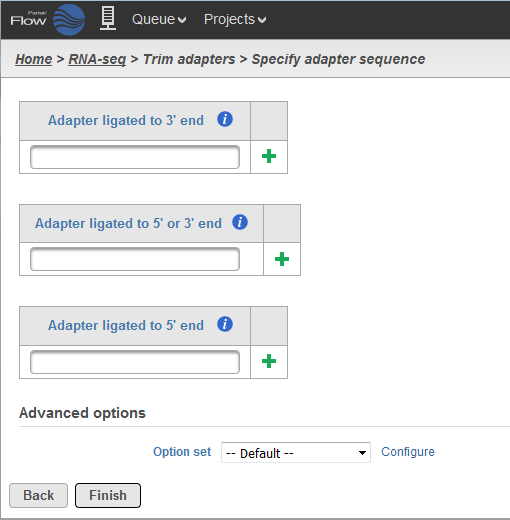
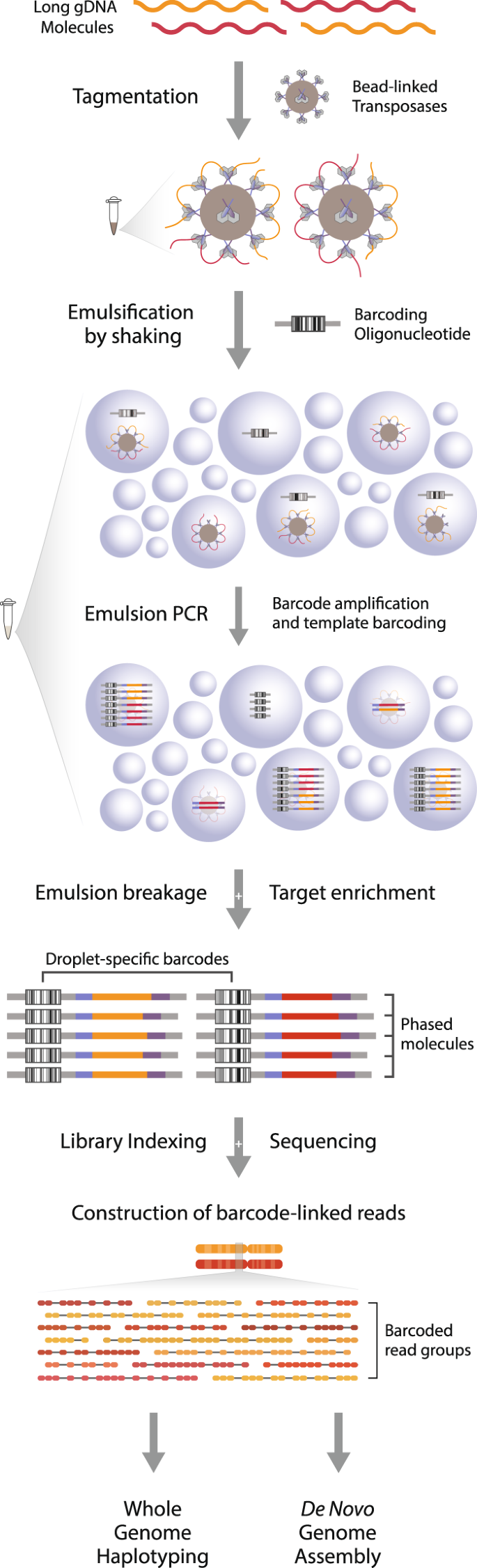
Cutadapt remoYes adapter sequences from high-throughput sequencing reads Introduction Implementation Features
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text

Assessing the utility of long-read nanopore sequencing for rapid and efficient characterization of mobile element insertions - Laboratory Investigation
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text